Bioinformatics: Where code meets biology
A brief introduction to the future of healthcare.
A brief introduction to the future of healthcare
You and I have 3 billion things in common. Or close to it anyway. And it’s not just you and I, everyone else is the same.
What could all these things be?
I like sports. Do you?
How about machine learning? I’m into that.
We could keep going and figure it out but it would take a while to get to 3 billion.
Even if we did make it there, we would’ve been comparing the wrong things.
Alright, well what are the right things?
Our DNA. Or more specifically, the nucleobases that combine to form the nucleic acids which chain together to build our DNA.
DNA is the language of life. It’s the code which sets the foundations for all living organisms.
Much like a book is comprised of some combination of the letters of the alphabet, you and I are comprised of some combination of around 3.2 billion nucleobases — adenine (A), thymine (T), cytosine (C) and guanine (G). The collective sequence is called a genome.
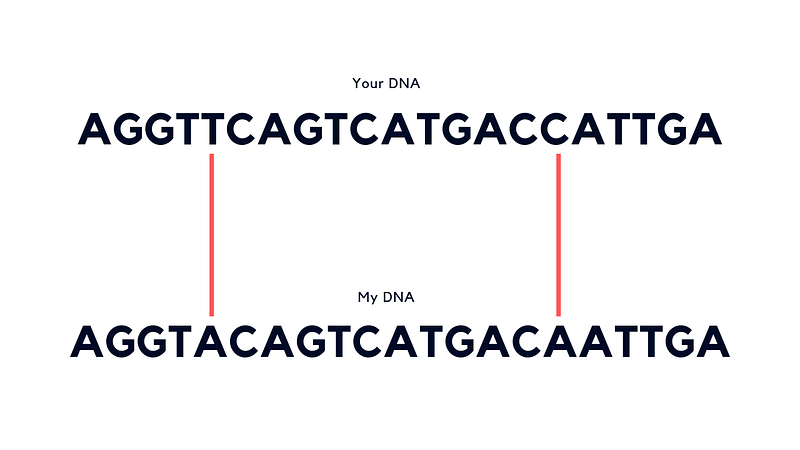
If we lined up our 3.2 billion or so A’s, C’s, G’s and T’s, 3 billion or so would match but 20–30 million would be different. Within those differences could explain why I’ve got blonde hair and you’ve got a different colour. And if you’re blonde too, we can add it to the list of things in common.
I’ve left out a few things here, such as, how different groups of letters encode for proteins, other groups make up genes, how genes have different versions within themselves and more.
Why?
Because DNA is a complex beast. Understanding how all 3.2 billion letters interact is still a large topic of research. And it’s been known for a while there’s no way a single person could interpret such a language.
This is where the power of computers comes in. It’s when the language of life meets the language of nature. And it’s where bioinformatics emerges.
Bioinformatics combines the principles of biology, computer science, mathematics, statistics to understand biology data.
Finding the Origin of Replication
The first example problem in the Coursera Bioinformatics Specialization involves finding the origin of replication. The origin of replication is a sequence of a genome where replication starts.
Before you were born, you started as a single cell and then one between two, two became four and eventually, four became you. For the first cell to divide, it had to replicate its genome. The same goes for each subsequent cell.
Knowing this, you can start to imagine how finding the origin of replication can be valuable.
Say there were a group of cells which had proteins within them which were very good at fighting cancer. How could we get more of these cells to better our defence?
One way might be to look for the replication of origin in our good cells, find it, and then use the information to produce more cells outside of the body and later put them back in.
Let’s try.
Storytime
Since your biologist friend knows you’ve been practising your coding skills, she comes to you and asks for help.

After many experiments of cutting portions of DNA out of the sample of the cell above and seeing the cell would replicate or not. She thinks she’s found the origin of replication.
To be sure, she wants to know how many times it occurs throughout the whole sequence and if it’s significant enough to be the actual origin of replication.
She shows you the files and you get to work.
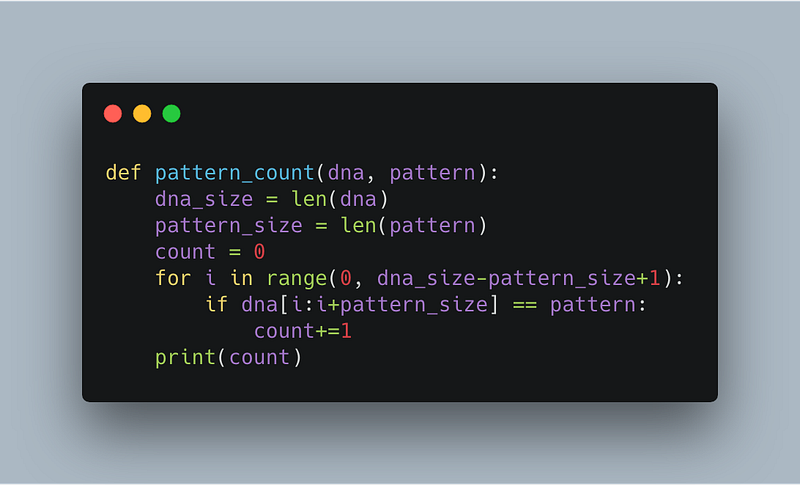
After a little tinkering, you think your code is ready to run. It goes through a DNA sequence and looks for the pattern if it finds a match, it updates a counter. When it reaches the end of the DNA pattern, it returns the count (the number of times the pattern occurred in the DNA sequence).
“What’s the pattern?” you ask.
“TGTAGTGTG.”
You help your friend run the code.
The result comes back as 18.
The pattern she found occurred 18 times throughout the DNA sequence of the good cells.
“Is that significant?” you ask.
“Well the probability of it occurring once is less than 0.004%, so 18 times must mean something but I’d have to check.”¹
You’re good at code but not so good at statistics, your biologist friend isn’t either. She goes back to the lab to find her statistician friend and run more tests.
What’s happened here is the combination of several fields. Your biologist friend found a potential replication of origin through experimentation but it was long and tedious. To help out, you offered some of your computer science skills. And then to find out whether your result was statistically significant.
This crossover of different fields is an example of bioinformatics at work. Each field brings insights to the table but putting them together makes them far more valuable.
The future of healthcare
The scenario above is a simplified example, bioinformatics in the real world takes far more steps.
First, how do you get a genome?
Well, luckily this part has seen rapid advancement over the past few decades. The first human genome took 23 years, teams all over the world and billions of dollars to sequence. Now you can get it done for under $15,000 in a few days.²
Okay, so you’ve got a genome, now what do you do?
Good question. This is where new methods of exploring DNA with different statistical and computational methods are actively being researched.
The sequence of DNA you saw above was linear, a single string of characters. But actual DNA is different, it’s comprised of two strands (one is the inverse opposite of the other) and different regions don’t always interact in the same way. So finding a pattern may not be as straight forward as what we’ve seen.
Well, say you did understand the genome a little more, how can it be applied?
The most interesting application of bioinformatics is personalised medicine. Healthcare tailored for an individual rather than an individual tailored to healthcare.
A project I’ve had the privilege to see unfold first hand is Immunotherapy Outcome Prediction or IOP by Max Kelsen. The goal is to use whole genome data to develop tools for predicting the outcome of immunotherapy treatments on patients with cancer. Some patients respond better to immunotherapy than others, why? Could the answer be hidden within their genomes?
Bioinformatics applications don’t stop at predicting cancer treatment outcomes either.
Nutrigenomics deals with the interaction of food and the genome. Think of a diet plan created specifically to work with your DNA.
My Dad takes a handful of drugs every day to aid with his Parkinson’s and Alzheimer’s symptoms. The first few years of taking them didn’t go well. It was only after a couple of rounds of trying different medications did he find some which didn’t leave him feeling worse. Could Pharmacogenomics, the creation of pharmaceuticals with regards to a person’s genome, help others find the right medication sooner?
Health and technology aren’t going away anytime soon. And we’re only scratching the surface with what’s possible at the intersection of these two fields.
My entry to bioinformatics only began a few days ago by starting the Bioinformatics Specialization on Coursera. But as I learn and experience more, I’ll be sure to share what I find. Be sure to follow along for more.
A video version of this article can be found on YouTube.
¹ To calculate the figure of 0.004%, the result of (1000–9+1)*(0.25)⁹ (probability of a certain pattern of A, C, G, T of length 9 occurring in a random sequence of A, C, G, T) was rounded to the nearest thousandth.
² Companies such as 23 and me perform genetic testing for far less, however, these services use genotyping rather than whole genome sequencing.
